Electromyography
Support for Electromyography (EMG) was developed as a BIDS Extension Proposal. Please see Citing BIDS on how to appropriately credit this extension when referring to it in the context of the academic literature.
Example datasets
Electromyography datasets formatted according to this specification are available on the BIDS examples repository and can be emulated when curating new datasets.
EMG data
Template:
sub-<label>/
[ses-<label>/]
emg/
sub-<label>[_ses-<label>]_task-<label>[_acq-<label>][_run-<index>][_recording-<label>]_emg.bdf
sub-<label>[_ses-<label>]_task-<label>[_acq-<label>][_run-<index>][_recording-<label>]_emg.edf
sub-<label>[_ses-<label>]_task-<label>[_acq-<label>][_run-<index>][_recording-<label>]_emg.json
sub-<label>[_ses-<label>]_task-<label>[_acq-<label>][_run-<index>][_recording-<label>]_events.json
sub-<label>[_ses-<label>]_task-<label>[_acq-<label>][_run-<index>][_recording-<label>]_events.tsv
Legend:
-
For more information about filename elements (for example, entities, suffixes, extensions), follow the links embedded in the filename template.
-
<matches>is a placeholder to denote an arbitrary (and valid) sequence of entities and labels at the beginning of the filename (only BIDS "raw"). -
<source-entities>is a placeholder to denote an arbitrary sequence of entities and labels at the beginning of the filename matching a source file from which the file derives (only BIDS-Derivatives). -
Filename entities or directories between square brackets (for example,
[_ses-<label>]) are OPTIONAL. -
Some entities may only allow specific values, in which case those values are listed in
<>, separated by|. -
_<suffix>means that there are several (>6) valid suffixes for this filename pattern. -
.<extension>means that there are several (>6) valid extensions for this file type. -
[.gz]means that both the unzipped and gzipped versions of the extension are valid.
EMG device manufacturers use a variety of formats for storing raw data, and there is no single standard that all researchers agree on. For BIDS, EMG data MUST be stored in one of the following formats:
| Format | Extension(s) | Description |
|---|---|---|
| Biosemi data format | .bdf |
Each recording consists of a single .bdf file. bdf+ files are permitted. The capital .BDF extension MUST NOT be used. |
| European data format | .edf |
Each recording consists of a single .edf file. edf+ files are permitted. The capital .EDF extension MUST NOT be used. |
EDF, EDF+, BDF, and BDF+ are all open data formats with broad support in various programming languages for reading and writing the files. BDF and BDF+ formats store data samples using 3 bytes instead of 2 bytes as in EDF and EDF+ formats, allowing for greater resolution. EDF+/BDF+ accommodate more header metadata than EDF/BDF, and support storing event or annotation information in the file. Thus it is RECOMMENDED to use the BDF+ data format.
The recording-<label> entity MAY be used to indicate
simultaneous acquisition of data from multiple EMG devices, in cases where the devices
store data in separate data files.
If separate devices are being used with separate sampling rates, start times, or other
acquisition parameters, the recording-<label> entity MUST be used to distinguish between them.
The synchronization of data from multiple devices SHOULD be described in the
Scans (scans.tsv) file, using the
acq_time entity.
In cases where the allowed formats do not encode all relevant metadata present in the device manufacturer's native file format, we encourage users to provide the additional metadata in the sidecar JSON file.
Terminology: Electrodes vs. Channels
For proper documentation of EMG recording metadata it is important to understand the difference between an electrode and a channel: an EMG electrode is a single point of electrical contact with the body, whereas a channel is the combination of the analog differential amplifier (which combines signals from two electrode sources) and the analog-to-digital converter, which results in a potential difference (voltage) that forms the actual data stored in the EMG dataset. We employ the following short definitions:
-
Electrode: A single point of contact between the acquisition system and the recording site (whether on the skin surface or intramuscular). Multiple electrodes can be organized as arrays, grids, leads, strips, probes, shafts, and so on.
-
Channel: A single analog-to-digital converter in the recording system that regularly samples the value of a transducer, which results in the signal being represented as a time series in the digitized data. This can be connected to two electrodes (to measure the potential difference between them), a magnetic field or magnetic gradient sensor, temperature sensor, accelerometer, and so forth.
This distinction is especially important for EMG because a common type of EMG device (often erroneously called a "bipolar electrode") comprises two electrodes at a fixed distance, wired such that the electrode pair is necessarily converted to a single channel by the amplifier (in other words, one of the electrodes necessarily acts as the reference for the other). In other kinds of devices, researchers may have control over which electrode(s) to use as the reference for each channel.
Note also that although the reference and ground electrodes are often referred to as channels, in most common EMG systems they are not recorded by themselves. Therefore they are usually not represented as channels in the data.
Describing sensor locations
Each EMG-BIDS dataset SHOULD include sensor location information that is as accurate as possible given the measurement means available to the researcher(s). Below are examples ranging from most accurate to least accurate, which can serve as a guide to where sensor placement information should be stored in the dataset.
-
Electrode locations and anatomical landmarks are digitized with a 3D localizer device such as Polhemus FasTrak or a 3D scanner. 3D coordinates for each electrode are given in
x,y,zcolumns of*_electrodes.tsv, and the coordinate system definition is given in*_coordsystem.json. -
Electrode locations are measured relative to nearby anatomical landmarks using a flexible measuring tape. 2D or 3D coordinates for each electrode are given in
x,y, and (optionally)zcolumns of*_electrodes.tsv, and the coordinate system definition in*_coordsystem.jsonspecifies the axis directions with reference to those anatomical landmarks. -
The arrangement of electrodes in a group or grid is measured (or known from the device manufacturer), and is provided in
x,y, and (optionally)zcolumns of*_electrodes.tsv, with a device-internal "child" coordinate system defined in a*_coordsystem.jsonfile. An anatomically-defined "parent" coordinate system is also defined in a different*_coordsystem.jsonfile, and the name and coordinates of one electrode (the "anchor" electrode) from each "child" group or grid is provided in the "parent" coordinate system. This allows the approximate anatomical locations of all electrodes to be calculated by treating the device-internal coordinates as offsets from the anchor coordinate. The two*_coordsystem.jsonfiles are distinguished by thespaceentity in their filenames; the labels used for that entity may be arbitrary (for example,*_space-grid_coordsystem.jsonfor the child coordinate system, and*_space-forearm_coordsystem.jsonfor the parent coordinate system). -
Individual electrode locations are chosen by visual inspection, palpation, or functional localizers. No measured coordinates are provided; placement information is given either in the
EMGPlacementSchemeDescriptionfield of*_emg.json(when the placement approach is the same for all sensors), or in theplacement_descriptioncolumn of*_channels.tsv(when the placement approach varied across sensors).
More details on each of these scenarios is given below in the Channels description, Electrodes description, and Coordinate system sections.
A fifth scenario unique to EMG involves location information that is fairly accurate
(as in scenarios (1) and (2) above) but it locates bipolar sensor devices rather than
electrodes.
In this case, researchers SHOULD provide *_electrodes.tsv even though information about
the individual electrode contacts is not available.
Rather, each entry from *_channels.tsv should have a corresponding row in *_electrodes.tsv
where the location information is given, and *_coordsystem.json should also be provided
(as in scenarios (1) or (2) above).
EMG in combination with other modalities
When EMG is recorded alongside other BIDS modalities (such as EEG or motion), researchers SHOULD define how recordings are synchronized. Options include:
-
Recording time stamps from a common clock source on the LATENCY channel of each modality’s acquisition device.
-
Recording experimental events (usually as TTL pulses) on dedicated channels of each modality’s acquisition devices.
-
Storing the acquisition time (relative to a common clock source) of the first data point of each modality’s recording in the acq_time column of the *_scans.tsv file. Note that the BIDS date time format allows optional fractional seconds, which SHOULD be used to maximize the precision of the synchronization.
-
Record the start- and stop-time of one modality’s recording (relative to the other modality’s clock) in the *_events.tsv file for the other modality.
It is up to the user to decide which approach is most appropriate given their experimental setup, and to identify which modality will serve as the 'main' clock source for synchronization purposes (or whether a separate external clock source is used).
Sidecar JSON (*_emg.json)
For consistency between studies and institutions, we encourage users to extract the values of metadata fields from the actual raw data. Whenever possible, please avoid using ad hoc wording.
These fields MUST be present:
| Key name | Requirement Level | Data type | Description |
|---|---|---|---|
| EMGPlacementScheme | REQUIRED | string | The way in which electrode placement locations were determined. If electrode locations are provided as coordinates in *_electrodes.tsv in reference to a coordinate system defined in *_coordsystem.json, then specify "Measured". If approach to placement varied across sensor devices, specify "ChannelSpecific" and include placement details in the placement_scheme column of *_channels.tsv. If a standardized electrode placement procedure (for example, SENIAM 8) was followed, or if an ad-hoc procedure was followed, specify "Other" and provide details in EMGPlacementSchemeDescription.Must be one of: "ChannelSpecific", "Measured", "Other". |
| EMGPlacementSchemeDescription | OPTIONAL, but REQUIRED if EMGPlacementScheme is "Other" |
string | Additional details of the approach to EMG sensor placement. Descriptions SHOULD NOT simply give the name of the targeted muscle (use the target_muscle column in *_channels.tsv for that), but rather SHOULD reflect the process used by the researcher(s) when placing electrodes. For example, EMG electrode sites may be chosen by measured distance from anatomical landmarks, by visual reference to (parts of) target muscles, by palpation of the skin to locate target muscles, or by functional localization (temporary electrode placement at several sites during prescribed behavior, until a site yielding strong EMG signal is found). |
| EMGReference | REQUIRED | string | Description of the approach to signal referencing. If the same signal source is used as reference for all channels (often called a "monopolar" recording), specify the signal source (typically an electrode name). If different channels have different reference electrodes, this field SHOULD contain the string "ChannelSpecific" and each channel's reference SHOULD be given in the reference column of *_channels.tsv. For bipolar recording devices (where a single sensor unit contains two rigidly separated contacts, and the paired contacts are not listed independently in *_electrodes.tsv), this field SHOULD be "Bipolar". |
| SamplingFrequency | REQUIRED | number | Sampling frequency (in Hz) of all the data in the recording, regardless of their type (for example, 2400). The sampling frequency of data channels that deviate from the main sampling frequency SHOULD be specified in the channels.tsv file. |
| PowerLineFrequency | REQUIRED | number or "n/a" |
Frequency (in Hz) of the power grid at the geographical location of the instrument (for example, 50 or 60). |
| RecordingType | REQUIRED | string | Defines whether the recording is "continuous", "discontinuous", or "epoched", where "epoched" is limited to time windows about events of interest (for example, stimulus presentations or subject responses).Must be one of: "continuous", "epoched", "discontinuous". |
| SoftwareFilters | REQUIRED | object of objects or "n/a" |
Object of temporal software filters applied, or "n/a" if the data is not available. Each key-value pair in the JSON object is a name of the filter and an object in which its parameters are defined as key-value pairs (for example, {"Anti-aliasing filter": {"half-amplitude cutoff (Hz)": 500, "Roll-off": "6dB/Octave"}}). |
Note the RecordingType, which depends on whether the data stream on disk is interrupted or not.
Continuous data is by definition 1 segment without interruption.
Epoched data consists of multiple segments that all have the same length
(for example, corresponding to trials) and that have gaps in between.
Discontinuous data consists of multiple segments of different length,
for example due to a pause in the acquisition.
These fields SHOULD be present:
| Key name | Requirement Level | Data type | Description |
|---|---|---|---|
| EMGChannelCount | RECOMMENDED | integer | Number of EMG channels. Must be a number greater than or equal to 0. |
| HardwareFilters | RECOMMENDED | object of objects or "n/a" |
Object of temporal hardware filters applied, or "n/a" if the data is not available. Each key-value pair in the JSON object is a name of the filter and an object in which its parameters are defined as key-value pairs. For example, {"Highpass RC filter": {"Half amplitude cutoff (Hz)": 0.0159, "Roll-off": "6dB/Octave"}}. |
| RecordingDuration | RECOMMENDED | number | Length of the recording in seconds (for example, 3600). |
These fields MAY be present:
| Key name | Requirement Level | Data type | Description |
|---|---|---|---|
| ElectrodeMaterial | OPTIONAL | string | Material of the electrode (for example, Tin, Ag/AgCl, Gold). If multiple sensor devices with heterogeneous electrode materials are recorded into the same data file, electrode material SHOULD be recorded in the material column of *_electrodes.tsv instead. If instead those devices were recorded into separate data files (distinguished for example by the recording entity), separate *_emg.json files (similarly distinguished) MAY be included, each with their own ElectrodeMaterial values. |
| ElectrodeType | OPTIONAL | string | Type of the electrode (for example, cup, ring, clip-on, wire, needle). |
| EMGGround | OPTIONAL | string | Description of the location of the ground electrode (for example, "right radial styloid process"). |
| EpochLength | OPTIONAL, but RECOMMENDED if RecordingType is "epoched" | number | Duration of individual epochs in seconds (for example, 1) in case of epoched data. If recording was continuous or discontinuous, leave out the field.Must be a number greater than or equal to 0. |
| Gain | OPTIONAL | number | Signal gain from an in-line amplifier, applied between the EMG sensor/device and the data acquisition computer. See also Preamplification. |
| InterelectrodeDistance | OPTIONAL | number | Distance between pairs or grids of electrodes rigidly attached to the same sensor device. If multiple sensor devices with heterogeneous inter-electrode distances are recorded into the same data file, inter-electrode distance SHOULD be recorded in the interelectrode_distance column of *_electrodes.tsv instead. If instead those devices were recorded into separate data files (distinguished for example by the recording entity), separate *_emg.json files (similarly distinguished) MAY be included, each with their own InterelectrodeDistance values. |
| Preamplification | OPTIONAL | number | Amplification built into an EMG bipolar sensor, electrode grid, or other device. See also Gain. |
| SkinPreparation | OPTIONAL | string | How skin was prepared prior to sensor device placement, for example, "alcohol wipe" or "abrasive gel". |
| SubjectArtefactDescription | OPTIONAL | string | Freeform description of the observed subject artifact and its possible cause (for example, "Vagus Nerve Stimulator", "non-removable implant"). If this field is set to "n/a", it will be interpreted as absence of major source of artifacts except cardiac and blinks. |
| TriggerChannelCount | OPTIONAL | integer | Number of channels for digital (binary TTL) triggers or analog equivalents (TTL in volt). Corresponds to the TRIG channel type.Must be a number greater than or equal to 0. |
Hardware information
| Key name | Requirement Level | Data type | Description |
|---|---|---|---|
| Manufacturer | RECOMMENDED | string | Manufacturer of the equipment that produced the measurements. |
| ManufacturersModelName | RECOMMENDED | string | Manufacturer's model name of the equipment that produced the measurements. |
| SoftwareVersions | RECOMMENDED | string | Manufacturer's designation of software version of the equipment that produced the measurements. |
| DeviceSerialNumber | RECOMMENDED | string | The serial number of the equipment that produced the measurements. A pseudonym can also be used to prevent the equipment from being identifiable, so long as each pseudonym is unique within the dataset. |
| ElectrodeManufacturer | RECOMMENDED | string | Can be used if all electrodes are of the same manufacturer (for example, "AD-TECH", "DIXI"). If electrodes of different manufacturers are used, please use the corresponding table in the _electrodes.tsv file. |
| ElectrodeManufacturersModelName | RECOMMENDED | string | If different electrode types are used, please use the corresponding table in the _electrodes.tsv file. |
Task information
| Key name | Requirement Level | Data type | Description |
|---|---|---|---|
| TaskName | REQUIRED | string | Name of the task. No two tasks should have the same name. The task label included in the filename MAY be derived from this "TaskName" field by removing all non-alphanumeric or + characters (that is, all except those matching [0-9a-zA-Z+]), and potentially replacing spaces with + to ease readability. For example "TaskName" "faces n-back" or "head nodding" could correspond to task labels faces+n+back or facesnback and head+nodding or headnodding, respectively. A RECOMMENDED convention is to name resting state task using labels beginning with rest. |
| TaskDescription | RECOMMENDED | string | Longer description of the task. |
| Instructions | RECOMMENDED | string | Text of the instructions given to participants before the recording. This is especially important in context of resting state recordings, to distinguish resting positions with different postures (for example, "seated", "reclined"). |
Note that the TaskName field does not have to be a "behavioral task" that subjects perform,
but can reflect some information about the conditions present when the data was acquired
(for example, "treatment", "control", or "sleep").
Institution information
| Key name | Requirement Level | Data type | Description |
|---|---|---|---|
| InstitutionName | RECOMMENDED | string | The name of the institution in charge of the equipment that produced the measurements. |
| InstitutionAddress | RECOMMENDED | string | The address of the institution in charge of the equipment that produced the measurements. |
| InstitutionalDepartmentName | RECOMMENDED | string | The department in the institution in charge of the equipment that produced the measurements. |
Example *_emg.json
{
"EMGChannelCount":4,
"EMGGround":"n/a",
"EMGPlacementScheme":"Other",
"EMGPlacementSchemeDescription":"midpoint between cubital fossa and radial styloid process",
"EMGReference":"4 reference electrodes built-in to device, placed near the midpoint between radial and ulnar styloid processes on the volar surface",
"HardwareFilters":{"Highpass RC filter": {"Half amplitude cutoff (Hz)": 0.0159, "Roll-off": "6dBOctave"}},
"InstitutionAddress":"9500 Gilman Drive 0559, La Jolla, CA 92093",
"InstitutionalDepartmentName":"Swartz Center for Computational Neuroscience",
"InstitutionName":"University of California San Diego",
"Instructions":"Jump straight upward as high as you can, while keeping your arms at your sides.",
"Manufacturer":"Delsys",
"ManufacturersModelName":"Trigno® Galileo",
"PowerLineFrequency":60,
"RecordingDuration":123.456,
"RecordingType":"continuous",
"SamplingFrequency":1000,
"SoftwareFilters":"n/a",
"TaskDescription":"jumping with stationary arms",
"TaskName":"jumping"
}
Channels description (*_channels.tsv)
Template:
sub-<label>/
[ses-<label>/]
emg/
sub-<label>[_ses-<label>]_task-<label>[_acq-<label>][_run-<index>][_recording-<label>]_channels.json
sub-<label>[_ses-<label>]_task-<label>[_acq-<label>][_run-<index>][_recording-<label>]_channels.tsv
Legend:
-
For more information about filename elements (for example, entities, suffixes, extensions), follow the links embedded in the filename template.
-
<matches>is a placeholder to denote an arbitrary (and valid) sequence of entities and labels at the beginning of the filename (only BIDS "raw"). -
<source-entities>is a placeholder to denote an arbitrary sequence of entities and labels at the beginning of the filename matching a source file from which the file derives (only BIDS-Derivatives). -
Filename entities or directories between square brackets (for example,
[_ses-<label>]) are OPTIONAL. -
Some entities may only allow specific values, in which case those values are listed in
<>, separated by|. -
_<suffix>means that there are several (>6) valid suffixes for this filename pattern. -
.<extension>means that there are several (>6) valid extensions for this file type. -
[.gz]means that both the unzipped and gzipped versions of the extension are valid.
A channel is represented by one time series recorded with the recording system.
Although this information can often be extracted from the EMG recording,
listing it in a simple .tsv document makes it easy to browse or search
(for example, searching for recordings with a sampling frequency of >=1000 Hz).
Hence, the *_channels.tsv file is RECOMMENDED.
Channels SHOULD appear in the table in the same order they do in the EMG data file.
Additional columns MAY be included to provide additional information about the channels.
Note that electrode positions SHOULD NOT be added to this file but to *_electrodes.tsv.
The columns of the channels description table stored in *_channels.tsv are:
| Column name | Requirement Level | Data type | Description |
|---|---|---|---|
| name | REQUIRED | string | Label of the channel. Values in name MUST be unique.This column must appear first in the file. |
| type | REQUIRED | string | Type of channel; MUST use the channel types listed below. Note that the type MUST be in upper-case. This column must appear second in the file. For a list of valid values for this column, see the associated glossary entry. |
| units | REQUIRED | string | Physical unit of the value represented in this channel, for example, V for Volt, or fT/cm for femto Tesla per centimeter (see Units). This column must appear third in the file. |
| description | OPTIONAL | string | Brief free-text description of the channel, or other information of interest. This column may appear anywhere in the file. |
| sampling_frequency | OPTIONAL | number | Sampling rate of the channel in Hz. This column may appear anywhere in the file. |
| signal_electrode | RECOMMENDED | string | The name of the electrode from which the reference's signal is subtracted to yield the channel's data. For channels originating from bipolar devices, the signal_electrode MAY be the same as the channel name. This column may appear anywhere in the file. |
| reference | RECOMMENDED | string | The reference for the given channel. When the reference is an electrode in *_electrodes.tsv, use the name of that electrode. For channels originating from bipolar devices, use "bipolar". For non-EMG channels (for example, trigger channels, microphones) use "n/a". This column may appear anywhere in the file. |
| group | RECOMMENDED | string or number | Which group the electrode or channel belongs to (typically this indicates electrodes or bipolar channels on the same grid, strip, or other device). This column may appear anywhere in the file. |
| target_muscle | RECOMMENDED | string | Name of the muscle(s) from which the EMG signal in this channel is believed to originate. When exact muscle is unknown (such as when using electrode grids that cover multiple muscles), groups of muscles may be named instead (for example "flexors of the left forearm"). This column may appear anywhere in the file. |
| placement_scheme | RECOMMENDED | string | A description of the electrode placement procedure used. See description of the EMGPlacementScheme field in *_emg.json. This column may appear anywhere in the file. Must be one of: "measured", "other". |
| placement_description | RECOMMENDED | string | Additional details of the approach to EMG sensor placement. See description of the EMGPlacementSchemeDescription field in *_emg.json. This column may appear anywhere in the file. |
| interelectrode_distance | RECOMMENDED | number | Distance between adjacent electrodes (in a pair or grid) that are rigidly attached to the same sensor device, or between the signal_electrode and its reference when each is independently placed. This column may appear anywhere in the file. |
| low_cutoff | OPTIONAL | number | Frequencies used for the high-pass filter applied to the channel in Hz. If no high-pass filter applied, use n/a. This column may appear anywhere in the file. |
| high_cutoff | OPTIONAL | number | Frequencies used for the low-pass filter applied to the channel in Hz. If no low-pass filter applied, use n/a. Note that hardware anti-aliasing in A/D conversion of all MEG/EEG/EMG electronics applies a low-pass filter; specify its frequency here if applicable. This column may appear anywhere in the file. Must be a number greater than or equal to 0. |
| notch | OPTIONAL | string | Frequencies used for the notch filter applied to the channel, in Hz. If notch filters are applied at multiple frequencies, these frequencies MAY be specified as a list, for example, [60, 120, 180]. If no notch filter was applied, use n/a. This column may appear anywhere in the file. |
| status | OPTIONAL | string | Data quality observed on the channel. A channel is considered bad if its data quality is compromised by excessive noise. If quality is unknown, then a value of n/a may be used. Description of noise type SHOULD be provided in [status_description]. This column may appear anywhere in the file. Must be one of: "good", "bad". |
| status_description | OPTIONAL | string | Freeform text description of noise or artifact affecting data quality on the channel. It is meant to explain why the channel was declared bad in the status column. This column may appear anywhere in the file. |
| Additional Columns | OPTIONAL | n/a |
Additional columns are allowed if they are defined in the associated metadata file. |
Restricted keyword list for field type in alphabetic order (shared with the MEG and EEG modality; however, only types that are common in EMG data are listed here). Note that upper-case is REQUIRED:
| Keyword | Description |
|---|---|
| ECG | ElectroCardioGram (heart) |
| EMG | ElectroMyoGram (muscle) |
| EOG | Generic EOG channel if HEOG or VEOG information not available |
| HEOG | Horizontal EOG |
| LATENCY | Latency of sample, in seconds from recording onset, typically from an external clock source. See acq_time column of the respective *_scans.tsv file. MUST be in form of s[.000000], where s reflects whole seconds, and .000000 reflects OPTIONAL fractional seconds. |
| MISC | Miscellaneous |
| REF | Reference channel |
| SYSCLOCK | Elapsed time since trial/recording start, as provided by the recording device. |
| TRIG | Analog (TTL in Volt) or digital (binary TTL) trigger channel |
| VEOG | Vertical EOG (electrooculogram) |
Examples of common free-form text for the description column (these should occur without enclosing quotation marks):
- n/a
- stimulus
- response
- skin conductance
- battery status
Example *_channels.tsv
See also the corresponding electrodes.tsv example.
name type units signal_electrode reference target_muscle
emg1 EMG V E1 E2 anterior belly of the digastric
emg2 EMG V E3 E4 levator angulis oris, zygomaticus major
emg3 EMG V E5 E6 platysma
emg4 EMG V E7 E8 obicularis oris
Electrodes description (*_electrodes.tsv)
Template:
sub-<label>/
[ses-<label>/]
emg/
sub-<label>[_ses-<label>][_task-<label>][_acq-<label>][_run-<index>][_recording-<label>]_electrodes.json
sub-<label>[_ses-<label>][_task-<label>][_acq-<label>][_run-<index>][_recording-<label>]_electrodes.tsv
Legend:
-
For more information about filename elements (for example, entities, suffixes, extensions), follow the links embedded in the filename template.
-
<matches>is a placeholder to denote an arbitrary (and valid) sequence of entities and labels at the beginning of the filename (only BIDS "raw"). -
<source-entities>is a placeholder to denote an arbitrary sequence of entities and labels at the beginning of the filename matching a source file from which the file derives (only BIDS-Derivatives). -
Filename entities or directories between square brackets (for example,
[_ses-<label>]) are OPTIONAL. -
Some entities may only allow specific values, in which case those values are listed in
<>, separated by|. -
_<suffix>means that there are several (>6) valid suffixes for this filename pattern. -
.<extension>means that there are several (>6) valid extensions for this file type. -
[.gz]means that both the unzipped and gzipped versions of the extension are valid.
File that gives the measured location, size, and other properties of EMG electrodes.
If an *_electrodes.tsv file is specified, a *_coordsystem.json file MUST be specified
as well.
Electrode locations may be specified in one of four ways:
-
Coordinates and landmarks digitized in situ. When 3D electrode locations are digitized in situ, the origin, orientation, and measurement unit of the coordinate system MUST be recorded in Cartesian coordinates according to the
EMGCoordinateSystemandEMGCoordinateUnitsfields in*_coordsystem.json, as described in the Coordinate Systems Appendix. In such cases,EMGCoordinateSystemSHOULD be specified asOther, theEMGCoordinateSystemDescriptionSHOULD contain a description of the origin and axis orientations of the 3D coordinate system, and thecoordinate_systemcolumn in*_electrodes.tsvmay be omitted. -
Measured coordinates in a single coordinate system defined by anatomical landmarks. This approach is suitable for individual electrodes placed close together on a single body part. In this case the
*_coordsystem.jsonfile MUST contain fieldsEMGCoordinateSystem,EMGCoordinateUnits, andEMGCoordinateSystemDescriptionas described in (1) above. For example, 4 electrodes all placed on the forearm (2 on the volar surface and 2 on the dorsal surface) could be located with coordinates in the following coordinate system:x: radial styloid process (RSP) → ulnar styloid process (USP);y: oleacranon process → cubital fossa;z: RSP-USP → lateral humerus epicondyle
It may be possible to omit the
zcolumn when all electrodes are on the same surface (in the above example, if all electrodes were on the volar surface and none were on the dorsal surface). Though it would be possible to omit bothzandyif all electrodes are colinear along the anatomically definedxaxis, theycolumn is required and thus researchers SHOULD enter0for all entries in columnyin such cases. In this case (of a single coordinate system for all electrodes) thecoordinate_systemcolumn in*_electrodes.tsvmay be omitted as in (1) above. -
Measured coordinates in multiple anatomically-defined coordinate systems. This approach is suitable for individual electrodes placed on multiple body parts. In this case there MUST be multiple
*_coordsystem.jsonfiles (distinguished by thespaceentity) for each coordinate system, each of which MUST contain the fieldsEMGCoordinateSystem,EMGCoordinateUnits, andEMGCoordinateSystemDescriptionas described in (1) above. Additionally, thecoordinate_systemcolumn in*_electrodes.tsvMUST indicate which named coordinate system the coordinate values in that row reflect, by referencing the label of thespaceentity for the corresponding*_coordsystem.jsonfile. For example,*_space-lowerLeg_coordsystem.jsonmay specify a coordinate system defined by lower leg landmarks for electrodes below the knee, and another called*_space-thigh_coordsystem.jsondefined by upper-leg landmarks for electrodes above the knee. In such a case, entries in thecoordinate_systemcolumn of*_electrodes.tsvMUST specify either"lowerLeg"or"thigh". As in (2) above, enter0for theycolumn values if all electrodes are colinear along the definedxaxis. -
Measured coordinates in "nested" coordinate systems. This approach is suitable for large electrode grids or similar devices. In this case, measured 2D locations in the
xandycolumns define the geometry of an electrode grid or group, relative to an arbitrary device-internal origin and axes (this is the "child" coordinate system, defined for example in a file*_space-dorsalGrid_coordsystem.json). The group's placement on the body is then described by selecting an "anchor" electrode and providing its coordinates within a "parent" coordinate system defined in reference to subject anatomy (as in (2) above), in a separate file such as*_space-forearm_coordsystem.json. Each coordinate system file MUST contain the fieldsEMGCoordinateSystem,EMGCoordinateUnits, andEMGCoordinateSystemDescriptionas described in (1) above. Additionally, each child coordinate system file MUST contain an entryParentCoordinateSystemgiving the value of thespaceentity for the file that defines the parent (anatomical) coordinate system, and entriesAnchorElectrodeandAnchorCoordinatesthat name an electrode and provide its coordinates in the parent coordinate system (respectively). For example, a coordinate system called "forearm" defined by forearm anatomical landmarks (as in (2) above) could be the parent coordinate system for two electrode grids placed on the volar and dorsal surfaces of the forearm. Each grid would have its own coordinate system file (named, for example,*_space-dorsalGrid_coordsystem.jsonand*_space-volarGrid_coordsystem.json) in which coordinates of each electrode are relative to a device-internal origin, such as the lower-leftmost electrode when the device is in a specified orientation. In such a scenario, entries in thecoordinate_systemcolumn of*_electrodes.tsvMUST specify either"volarGrid"or"dorsalGrid"for electrodes within each grid (electrodes external to the grid, such as separate reference or ground electrodes, MAY specify a different value in thecoordinate_systemcolumn: typically this will be the parent coordinate system or a different anatomically defined coordinate system as in (2) above). Note that the parent coordinate system may be defined in different units than the child coordinate system; for example, if an anchor electrode was placed 50% of the distance between the ulnar styloid process and cubital fossa, the unit of the parent coordinate system is"percent"andAnchorCoordinateswould have a value of50for thexcoordinate in*_space-volarGrid_coordsystem.json. Meanwhile, thexcolumn of*_electrodes.tsvgives the device-internal coordinates of the grid electrodes, which may be in a different unit such as"mm". See thecoordsystem.jsonsection for further details.
For details of how to specify the coordinate systems in each of these cases, see the
coordsystem.json section.
Required columns
The order of the required columns in the *_electrodes.tsv file MUST be as listed below.
| Column name | Requirement Level | Data type | Description |
|---|---|---|---|
| name | REQUIRED | string | Name of the electrode contact point. The combination of name and group MUST be unique.This column must appear first in the file. |
| x | REQUIRED | number | Recorded position along the x-axis. This column must appear second in the file. |
| y | REQUIRED | number | Recorded position along the y-axis. This column must appear third in the file. |
| z | OPTIONAL | number | Recorded position along the z-axis. This column must appear fourth in the file. |
| coordinate_system | RECOMMENDED | string | Name of the coordinate system defined in coordsystem.json in which the electrode's x, y, and (optionally) z coordinates are given. This column must appear fifth in the file. |
| type | RECOMMENDED | string | Type of the electrode (for example, cup, ring, clip-on, wire, needle). This column may appear anywhere in the file. |
| material | RECOMMENDED | string | Material of the electrode (for example, Tin, Ag/AgCl, Gold). This column may appear anywhere in the file. |
| impedance | RECOMMENDED | number | Impedance of the electrode, units MUST be in kOhm. This column may appear anywhere in the file. |
| group | RECOMMENDED | string or number | Which group the electrode or channel belongs to (typically this indicates electrodes or bipolar channels on the same grid, strip, or other device). The combination of name and group MUST be unique.This column may appear anywhere in the file. |
| Additional Columns | OPTIONAL | n/a |
Additional columns are allowed if they are defined in the associated metadata file. |
The recording-<label> entity MUST be used to indicate
simultaneous acquisition of data from multiple EMG devices, in cases where the devices
store data in separate data files.
For example:
└─ sub-01/
└─ ses-01/
└─ emg/
├─ sub-01_ses-01_recording-RectusFemoris_electrodes.tsv
├─ sub-01_ses-01_recording-VastusLateralis_electrodes.tsv
└─ sub-01_ses-01_recording-VastusMedialis_electrodes.tsv
Above, acquisitions are labeled with the target muscle, but other naming schemes are also
appropriate. For example, if bipolar and grid EMG devices are used simultaneously,
acquisitions may be labeled "bipolar" and "grid", or use the device manufacturer names.
For cases of multiple devices recording simultaneously into the same data file,
the group column MUST be used to distinguish which electrodes belong to which device.
Except in cases of simultaneous acquisitions from different devices into different data
files (as mentioned above), *_electrodes.tsv files SHOULD NOT be duplicated for each data file,
for example, during multiple runs of a task.
The inheritance principle MUST
be used to find the appropriate electrode positions for a given data file.
If electrodes are repositioned, it is RECOMMENDED to use multiple sessions to indicate this.
Example *_electrodes.tsv
Example *_electrodes.tsv for a 2 by 2 grid of EMG electrodes with 2.5 mm electrode diameter
and 10 mm inter-electrode distance.
name x y coordinate_system group diameter
001 0.0 0.0 VolarForearm grid1 2.5
002 10.0 0.0 VolarForearm grid1 2.5
003 0.0 10.0 VolarForearm grid1 2.5
004 10.0 10.0 VolarForearm grid1 2.5
001 0.0 0.0 DorsalForearm grid2 2.5
002 10.0 0.0 DorsalForearm grid2 2.5
003 0.0 10.0 DorsalForearm grid2 2.5
004 10.0 10.0 DorsalForearm grid2 2.5
Coordinate System JSON (*_coordsystem.json)
Template:
sub-<label>/
[ses-<label>/]
emg/
sub-<label>[_ses-<label>][_task-<label>][_acq-<label>][_space-<label>][_recording-<label>]_coordsystem.json
Legend:
-
For more information about filename elements (for example, entities, suffixes, extensions), follow the links embedded in the filename template.
-
<matches>is a placeholder to denote an arbitrary (and valid) sequence of entities and labels at the beginning of the filename (only BIDS "raw"). -
<source-entities>is a placeholder to denote an arbitrary sequence of entities and labels at the beginning of the filename matching a source file from which the file derives (only BIDS-Derivatives). -
Filename entities or directories between square brackets (for example,
[_ses-<label>]) are OPTIONAL. -
Some entities may only allow specific values, in which case those values are listed in
<>, separated by|. -
_<suffix>means that there are several (>6) valid suffixes for this filename pattern. -
.<extension>means that there are several (>6) valid extensions for this file type. -
[.gz]means that both the unzipped and gzipped versions of the extension are valid.
This *_coordsystem.json file contains the coordinate system in which electrode positions
are expressed.
Associated photos can also be provided.
The *_coordsystem.json is REQUIRED if the optional *_electrodes.tsv is specified;
please see the electrodes description section for
important guidance regarding how to specify electrode location.
Fields relating to the EMG coordinate system(s):
| Key name | Requirement Level | Data type | Description |
|---|---|---|---|
| EMGCoordinateSystem | REQUIRED | string | Defines the coordinate system for the EMG sensors. See the Coordinate Systems Appendix for a list of restricted keywords for coordinate systems. If "Other", provide definition of the coordinate system in EMGCoordinateSystemDescription. "Other" is the only valid value until standardized names become available for EMG coordinate systems.Must be one of: "Other". |
| EMGCoordinateUnits | REQUIRED | string | Units of the coordinates of EMGCoordinateSystem.Must be one of: "m", "mm", "cm", "percent", "n/a". |
| EMGCoordinateSystemDescription | RECOMMENDED, but REQUIRED if EMGCoordinateSystem is "Other" |
string | Free-form text description of the coordinate system, typically defining the origin and positive x,y,z axis directions, relative to anatomical landmarks. May also include a link to a documentation page or paper describing the system in greater detail. |
| ParentCoordinateSystem | OPTIONAL | string | The name of a JSON mapping key that defines the parent (anatomical) coordinate system in which this child (device) coordinate system is anchored. |
| AnchorCoordinates | OPTIONAL, but REQUIRED if ParentCoordinateSystem is present |
array of numbers | Coordinates of the AnchorElectrode in the parent (anatomical) coordinate system. This should be an array of numbers of length 3, in the units given in EMGCoordinateSystemUnits within the JSON object named in ParentCoordinateSystem. |
| AnchorElectrode | OPTIONAL, but REQUIRED if ParentCoordinateSystem is present |
string | Name of the electrode in the child (device) coordinate system whose coordinates in the parent (anatomical) coordinate system are given in AnchorCoordinates. |
The recording-<label> entity MUST be used to indicate
simultaneous acquisition of data from multiple EMG devices, in cases where the devices
store data in separate data files.
Except in cases of simultaneous acquisitions from different devices into different data
files (as just mentioned), *_coordsystem.json files SHOULD NOT be duplicated for each data file,
for example, during multiple runs of a task.
However, multiple *_coordsystem.json files distinguished by the space entity MUST be
used to describe separate anatomically-defined coordinate systems (for example, to situate
devices in reference to landmarks on different limbs).
Multiple *_coordsystem.json files distinguished by the space entity MUST also be used to describe
nested coordinate systems that give the relative positions of groups or grids of electrodes
mounted to a single device and situate that "child" coordinate system within an anatomically-defined
"parent" coordinate system that situates the placement multi-electrode device on the body.
The inheritance principle MUST
be used to find the appropriate coordinate system description for a given data file.
Example 1: *_coordsystem.json for digitized electrode positions
Here, a coordinate system is defined based on digitized cranial landmarks, and is suitable for EMG electrodes on the face whose locations are digitized with the same equipment as the cranial landmarks:
{
"EMGCoordinateSystem": "Other",
"EMGCoordinateSystemDescription": "x: left helix-tragus junction (LHJ) → right helix-tragus junction (RHJ); y: inion → nasion; z: midpoint between mastoid processes → vertex",
"EMGCoordinateSystemUnits": "m"
}
Example 2: *_coordsystem.json for measured electrode positions in a single coordinate system defined by anatomical landmarks
Here, a coordinate system is defined based on percentage of distances between anatomical landmarks of the forearm, and is suitable for localizing individual electrodes on the same body part:
{
"EMGCoordinateSystem": "Other",
"EMGCoordinateSystemDescription": "x: radial styloid process (RSP) → ulnar styloid process (USP); y: oleacranon process → cubital fossa; z: RSP-USP → lateral humerus epicondyle",
"EMGCoordinateSystemUnits": "percent"
}
Example 3: multiple *_coordsystem.json files for measured electrode positions in multiple anatomically-defined coordinate systems
Here, separate anatomically-based coordinate systems are defined for electrodes above and
below the elbow joint, and each is given a unique name ("Forearm" or "Humerus") which can
be referenced in the coordinate_system column of *_electrodes.tsv:
{
"EMGCoordinateSystem": "Other",
"EMGCoordinateSystemDescription": "x: radial styloid process (RSP) → ulnar styloid process (USP); y: oleacranon process → cubital fossa; z: RSP-USP → lateral humerus epicondyle",
"EMGCoordinateSystemUnits": "percent"
}
{
"EMGCoordinateSystem": "Other",
"EMGCoordinateSystemDescription": "x: medial humerus epicondyle (MHE) → lateral humerus epicondyle (LHE), y: oleacranon process → cubital fossa, z: MHE-LHE → greater humerus tubercule",
"EMGCoordinateSystemUnits": "percent"
}
Example 4: multiple *_coordsystem.json files for nested/anchored coordinate systems
Here, coordinate systems for the relative locations of electrodes in grids or groups are given in "child" coordinate systems ("BicepGrid", "VolarForearmGrid", "DorsalForearmGrid"), each of which is anchored to a "parent" coordinate system that is defined anatomically:
{
"EMGCoordinateSystem": "Other",
"EMGCoordinateSystemDescription": "x-axis left → right, y-axis bottom → top, when grid is oriented with leads at the bottom",
"EMGCoordinateSystemUnits": "mm",
"ParentCoordinateSystem": "Humerus",
"AnchorCoordinates": [40, 70, 0],
"AnchorElectrode": "E1"
}
{
"EMGCoordinateSystem": "Other",
"EMGCoordinateSystemDescription": "x-axis left → right, y-axis bottom → top, when grid is oriented with leads at the bottom",
"EMGCoordinateSystemUnits": "mm",
"ParentCoordinateSystem": "Forearm",
"AnchorCoordinates": [25, 50, 10],
"AnchorElectrode": "E1"
}
{
"EMGCoordinateSystem": "Other",
"EMGCoordinateSystemDescription": "x-axis left → right, y-axis bottom → top, when grid is oriented with leads at the bottom",
"EMGCoordinateSystemUnits": "mm",
"ParentCoordinateSystem": "Forearm",
"AnchorCoordinates": [75, 0, 90],
"AnchorElectrode": "E1",
}
{
"EMGCoordinateSystem": "Other",
"EMGCoordinateSystemDescription": "x: radial styloid process (RSP) → ulnar styloid process (USP); y: oleacranon process → cubital fossa; z: RSP-USP → lateral humerus epicondyle",
"EMGCoordinateSystemUnits": "percent"
}
{
"EMGCoordinateSystem": "Other",
"EMGCoordinateSystemDescription": "x: medial humerus epicondyle (MHE) → lateral humerus epicondyle (LHE), y: oleacranon process → cubital fossa, z: MHE-LHE → greater humerus tubercule",
"EMGCoordinateSystemUnits": "percent"
}
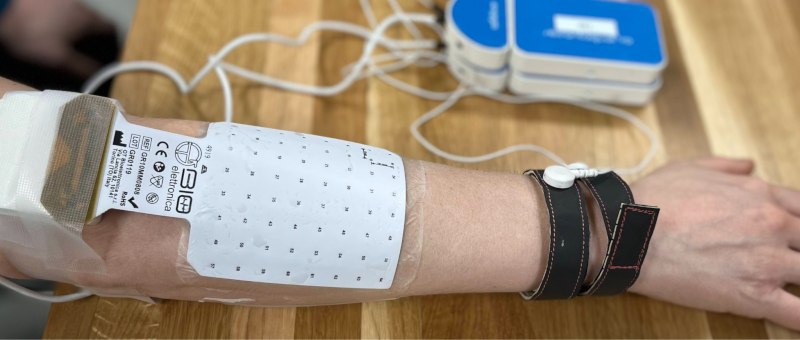
Photos of the electrode positions (*_photo.<extension>)
Template:
sub-<label>/
[ses-<label>/]
emg/
sub-<label>[_ses-<label>][_acq-<label>][_recording-<label>]_photo.jpg
sub-<label>[_ses-<label>][_acq-<label>][_recording-<label>]_photo.png
sub-<label>[_ses-<label>][_acq-<label>][_recording-<label>]_photo.tif
Legend:
-
For more information about filename elements (for example, entities, suffixes, extensions), follow the links embedded in the filename template.
-
<matches>is a placeholder to denote an arbitrary (and valid) sequence of entities and labels at the beginning of the filename (only BIDS "raw"). -
<source-entities>is a placeholder to denote an arbitrary sequence of entities and labels at the beginning of the filename matching a source file from which the file derives (only BIDS-Derivatives). -
Filename entities or directories between square brackets (for example,
[_ses-<label>]) are OPTIONAL. -
Some entities may only allow specific values, in which case those values are listed in
<>, separated by|. -
_<suffix>means that there are several (>6) valid suffixes for this filename pattern. -
.<extension>means that there are several (>6) valid extensions for this file type. -
[.gz]means that both the unzipped and gzipped versions of the extension are valid.
Photos of the electrode locations are OPTIONAL. Photos SHOULD include sufficient surrounding context to distinguish anatomical location. For example, photographs of electrodes on the limbs should include at least one adjacent joint. Photos may need to be cropped and/or blurred to conceal identifying features or entirely omitted prior to dataset distribution, depending on obtained consent.
If there are photos of the electrodes, the recording-<label> entity
MAY be specified, if separate photos are provided of each EMG device and the devices recorded
into separate data files.
The ses-<label> entity may be used to specify when the photo was taken.
Example *_photo.<extension>
└─ sub-01/
└─ ses-01/
├─ sub-01_ses-01_recording-DorsalForearmGrid_photo.jpg
└─ ...